Project leader: Professor Veronica Ciocanel
Project manager: Ray Tung
Team members: Maddie Dawson, Carson Dudley, Sasamon Omoma
This project has produced the following paper:
Madeleine Dawson, Carson Dudley, Sasamon Omoma, Hwai-Ray Tung and Maria-Veronica Ciocanel, Characterizing emerging features in cell dynamics using topological data analysis methods, Mathematical Biosciences and Engineering volume 20, issue 2, 3023-3046.
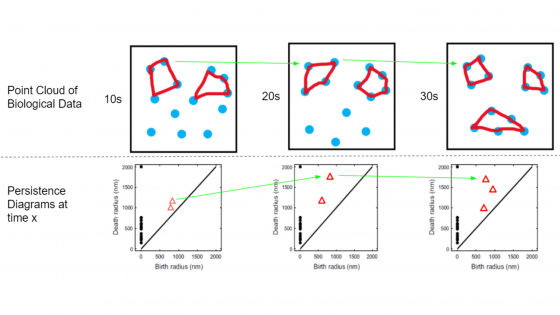
Actin is one of the most abundant proteins in cells which assembles to form polymer filaments. When actin filaments interact with motor proteins, they can organize into ring channels. These ring channels are present in many critical biological processes, such as cellular division, development, and wound healing. The onset of ring channels and their maintenance throughout time is not well understood. Our research proposes new and improved methods of analyzing ring channel dynamics throughout time. To accomplish this, we used topological data analysis (TDA), a growing branch of mathematics, to understand the shape of data. We successfully developed and applied TDA methods to synthetic and experimental time-series contractile ring data.
Our goal was to accurately match birth-death pairs between the persistence diagrams corresponding to protein organization in each time frame. We generated ‘paths’ of persistent features over time, which match the behaviors and geometry of the in silico and in vitro time-series data. We accomplished this by exploring several steps on the standard TDA pipeline. One objective was to encode the data generated by MEDYAN, a software that simulates actin filament interactions: we encoded the filamentous structure of the point cloud by modifying its distance matrix, which yielded a smoother, more continuous path of persistent features. We matched persistence diagrams over time using the Bottleneck and Wasserstein distance metrics. We concluded that these measures produced representative paths of the ring channel dynamics that improve on previous methods, and explored their limitations. We also investigated different filtration methods to generate the persistence diagram: Vietoris-Rips, flooding, and superlevel set filtration. Finally, we determined significance thresholds for separating signal and noise in persistence diagrams using Poisson Point Processes on the simulation domains. Our developed TDA methods successfully tracked multiple ring channels in actin organization throughout time in many biological contexts.